Blog | Aug. 30, 2023
MRD in AML: the next step in fighting leukemia

Biopsy is the removal of tissue from a living organism for microscopic examination and diagnosis. An Arab physician, Abulcasis, (1013–1107) is credited with performing one of the earliest diagnostic biopsies when he used a needle to puncture a goiter and then characterized the material.1 A French dermatologist Ernest Besnier introduced the word biopsie in 1879.2 It is a Greek-derived word (bio-life; opsia-to see) loosely translated as “view of the living.”
Tissue biopsy is the current standard-of-care in clinical cancer management and is the primary method for a definitive cancer diagnosis in solid tumors. Biopsies are used to: • identify the type of cancer,
• gene expression,
• and presence of treatment-resistant mutations.
Biopsies provide information about:
• tumor histology,
• presence of actionable biomarkers for subtyping,
• therapy selection, and
• molecular profiling for prediction and prognostication.3
While biopsies provide actionable information, utilization is hampered by the invasiveness, tumor location, tissue amount available, and accessibility of the primary or metastasized tumor for surgical removal. Utilizing repeated conventional biopsies to track tumor evolution is tedious and painful and may lead to complications, e.g., bleeding and/or the dislodging of cancer cells from the tumor, thus allowing them to spread elsewhere called cancer cell seeding.4 Moreover, the tumor genetic information is limited to the biopsy area which may be misinterpreted due to the tumor heterogeneity and multiple tumor sites. Also, serial solid biopsies are not practical for long-term monitoring of the patient.5
A tissue biopsy reflects the molecular composition of the tumor at the time the sample is taken; however, malignant neoplasms evolve continuously allowing cancers to adapt to changing environments, surviving treatments, leaving residual disease, and potential variability between the primary and metastasized tumor.4
Due to the limitations of tissue biopsy, detection of cancer through non-invasive techniques has been under investigation for more than two decades.6 Liquid biopsy is a minimally invasive test requiring a small sample of blood or urine that can detect cancer cells or genetic material that primary and/or metastatic lesions release into the body fluids.7 Tumor cells in circulation were first discovered in the late 19th century. The term “cell-free DNA (cfDNA)” was coined by Mandel and Metais in 1948 who detected cfDNA in the bloodstream of cancer patients.4 In 1977, another group detected larger amounts of cfDNA in cancer patients than healthy patients.8 Stroun et al. then showed that some plasma cfDNA was derived from circulating tumor DNA (ctDNA) and carried its molecular signature.9
Liquid biopsies have the potential to address key areas related to:
• diagnosis,
• prognosis, and
• therapeutics, i.e., monitoring the spread of cancer to other parts of the body, determining presence of genetic changes and/or mutations, evaluating efficacy of treatments, and emergence of resistance.10
Furthermore, serial liquid biopsies can monitor disease progression and perform surveillance to better inform therapeutic decisions for personalized medicine. The technology can provide a more comprehensive picture of a patient’s pathology and overcome the spatial limitations of a tissue biopsy taken from a single lesion within a single anatomic site.11
Liquid biopsies have the potential to be used for cancer screening and early detection of cancer. If detected early, cancer treatment is more effective, local tumors may be excised by surgery, and survival rates tend to be higher.12 However, prior to clinical adoption, liquid biopsy tests need to be clinically validated and assessed for reliability, reproducibility, and cost effectiveness. Liquid biopsies are best used for screening, identifying mutations in metastatic cancer, and tracking genetic changes to inform treatment.13
Late diagnosis reduces the chances of survival of cancer patients.14 Using ctDNA for early cancer diagnosis remains challenging due to the low amount of tumor DNA released into the circulation and its dilution with non-tumor DNA. With technologies such as qPCR or conventional NGS, the sensitivity of mutation detection is generally higher than 2-5% which is inadequate.15
However, newer technologies, such as droplet-based digital PCR (ddPCR) and optimized NGS, display greatly improved sensitivity, specificity, and precision for the quantitative detection of rare sequences in early-stage cancer with a sensitivity below 0.001%.16,17
ddPCR uses limiting dilutions in microtiter plate-based technology to partition individual target sequences from a complex mixture into separate compartments, allowing rare event to be detected at the level of single molecule. Aqueous droplets with volumes ranging from few femtoliters to nanoliters are dispersed in oil for compartmentalization of PCR reactions.18 Each droplet ideally contains no more than one haploid genome with one target mutant sequence and one wild-type allele labeled with different fluorophores and has a detection sensitivity below 0.001%.16
BEAMing (beads, emulsion, amplification, and magnetics, commercialized by Sysmex Inostics as OncoBEAM) was the first high-throughput ddPCR system described in 2003 for the detection and enumeration of genetic variants with a detection limit of one mutant DNA molecule in a background of 10,000 wild-type molecules.19
On the other hand, optimized NGS technology analyzes millions of ctDNA molecules at the same time and the sequence data are then aligned against a reference genome identifying genetic or epigenetic changes. Conventional NGS technologies such as Ion AmpliSeq, with detection sensitivity higher than 2%, are not suitable for the detection of rare mutations in ctDNA.20 Newer NGS technologies such as Plasma-Safe-SeqS (PSS) have greatly improved detection sensitivity and specificity of ctDNA.17,21
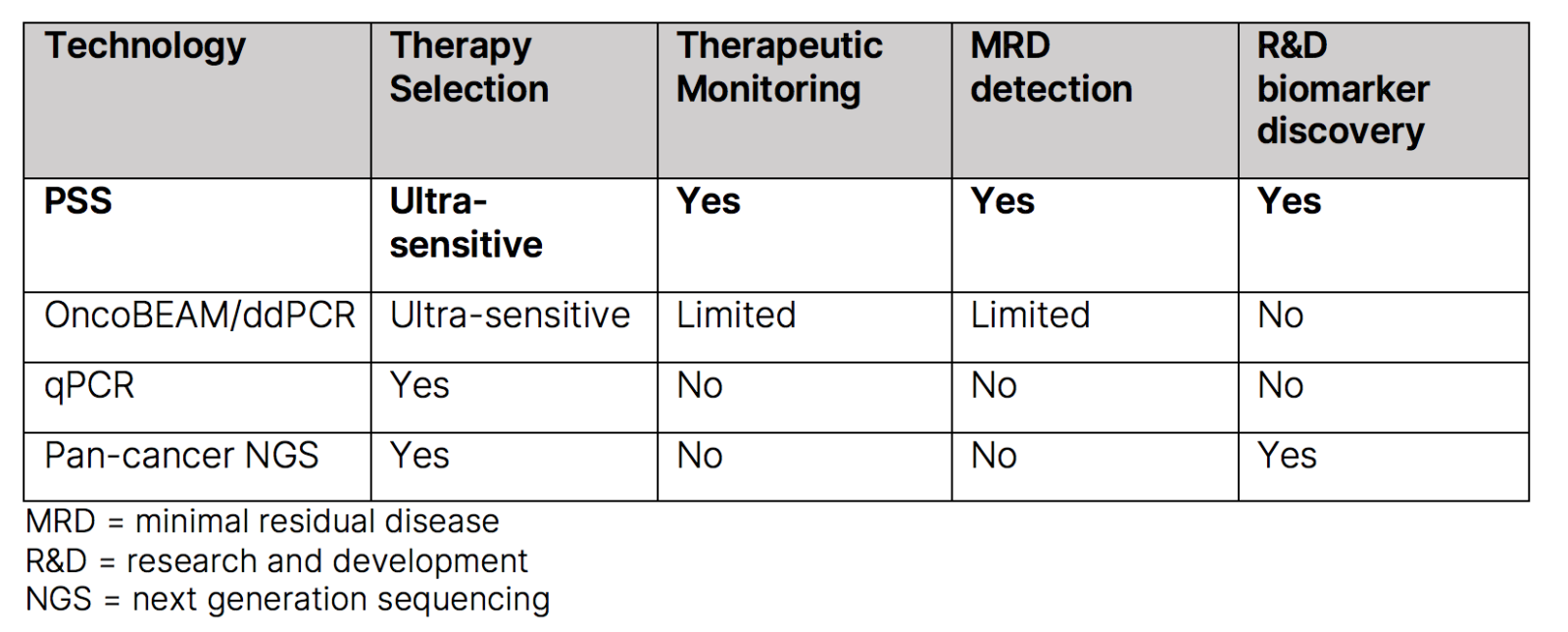
Compared to NGS, ddPCR experiments have easier set-up, are faster, present higher sensitivity, and do not require complex informatics support for analysis. However, it requires prior knowledge of genetic or epigenetic changes to be detected and present limited multiplex abilities.22 In contrast, NGS could identify novel genetic or epigenetic modifications and present high multiplexing capabilities but is time consuming and requires informatic support. Due to their varying strengths, many strategies have thus combined the use of NGS and ddPCR for liquid biopsy analysis.23
PSS is a low error rate technology, where each DNA molecule is assigned a unique identifier (UID), and UID families are amplified and deep sequenced. The platform discriminates real mutations from errors potentially introduced during the amplification and sequencing process, resulting in more accurate mutation detection. Assays using PSS technology are designed to preserve mutant molecules throughout the workflow, leaving a more robust sample to identify patients even with extremely low-level ctDNA. Other assays, particularly broad, hybrid-capture-based pan-cancer panels, may lose up to 40% of all input DNA during sample prep, which decreases reliable detection of ctDNA.24
With traditional broad panel NGS testing, sample-input DNA often limits assay sensitivity and results in increased costs since fixed pan-cancer NGS assays expend sequencing power on unnecessary gene regions.
By accurately identifying ctDNA across various tumor types, PSS’ best-in-class sensitivity can expedite and improve advanced therapeutic clinical development. PSS demonstrates equivalent performance to OncoBEAM digital PCR and is 10x more sensitive than other liquid biopsy NGS methods.25 Using a computational pipeline that performs barcode-mediated background error suppression and maximizes molecule retention, the technology can achieve detection sensitivity of 0.001% and specificity of 96%.26
PSS level of sensitivity allows clinical trial sponsors to accelerate trial enrollment and evaluate biomarker hypotheses with greater power.
References: